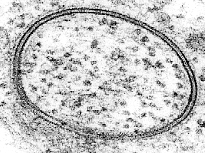
缝隙连接
编辑缝隙连接(间隙连接)是多种动物细胞类型之间的一种特殊细胞间连接。[1][2][3] 它们将两个细胞的细胞质直接相连,使各种分子、离子和电脉冲直接穿过细胞间的调节通道。[4][5]
一个缝隙连接通道由两个连接体(或半通道)组成,它们横跨细胞缝隙连接。[4][5] 缝隙连接类似于连接植物细胞的胞间连丝。
除了成年体完全发育的骨骼肌和移动类型细胞,如精子或红细胞,缝隙连接几乎存在于在机体的所有组织中。然而,在海绵和黏菌等简单生物中缝隙连接并未被发现。
缝隙连接也可以称为连接或黄斑通信。(虽然大多数神经组织无缝隙连接存在,但当在神经元或神经中也发现了被称为电突触的连接,就像牙髓中的神经细胞一样。)虽然神经元间接触与缝隙连接有一些相似之处,但根据现代定义,两者是不同的。
目录编辑
1 结构编辑
脊椎动物的缝隙连接半通道的连接蛋白主要是同源或异源六聚体。无脊椎动物缝隙连接是由无脊椎动物连接蛋白家族蛋白质组成。无脊椎动物连接蛋白蛋白与连接蛋白没有明显的序列同源性。[6]虽然同连接蛋白的序列不同,但无脊椎动物连接蛋白与脊椎动物的连接蛋白非常相似,表明无脊椎动物连接蛋白在体内形成缝隙连接的方式与脊椎动物的连接蛋白相同。[7][8][9]最近被鉴定的泛连接蛋白家族,[10] 最初被认为形成细胞间通道的一种蛋白(氨基酸序列与无脊椎动物连接蛋白相似),[11]实际上起着与细胞外环境相通的单膜通道的作用,已经被证明能传递钙离子和三磷酸腺苷(ATP)。[12]
在缝隙连接处,细胞间隙在2-4纳米之间,[13] 且每个细胞膜中的单位连接彼此对齐。[13]
由两个相同的半通道形成的缝隙连接称为同型通道,而具有不同半通道的缝隙连接称为异型通道。反过来,具有相同连接蛋白组成的半通道被称为同聚体,而具有不同连接蛋白的半通道是异聚体。通道的组成被认为影响缝隙连接的功能。
在无脊椎动物连接蛋白和泛连接蛋白进行充分表征前,根据基因定位和序列相似性,将编码连接蛋白缝隙连接的基因分为三组:甲、乙和丙(例如GJA1、GJC1)。[14][15][16]然而,连接蛋白基因不直接编码缝隙连接通道的表达;基因只能产生构成缝隙连接通道的蛋白质。基于该蛋白质分子量的另一种命名系统也很流行(例如:连接蛋白43 = GJA1,连接蛋白30.3 = GJB4)。
2 组织层次编辑
- DNA转化为连接蛋白的RNA。
- 一种连接蛋白有四个跨膜结构域
- 6连接蛋白创造一个连接子(半通道)。当不同的连接蛋白结合在一起形成一个连接子时,它被称为异聚连接子。
- 跨膜连接在一起的两个半通道构成缝隙连接通道。当两个相同的连接在一起形成一个缝隙连接通道时,它被称为同型GJ通道。当一个同聚连接子和一个异聚连接子结合在一起时,它被称为异型缝隙连接通道。当两个异聚连接子连接时,它也被称为异型缝隙连接通道。
- 几个缝隙连接通道(数百个)组装在一个被称为缝隙连接斑块的大分子复合体中。
3 连接子通道对的属性编辑
连接子通道对:
- 允许细胞之间的直接进行电信号传导,不同的连接蛋白亚单位可以赋予不同的单通道电导,范围从大约30 pS到500 pS。
- 允许细胞之间进行化学通信,一些第二信使分子,例如肌醇三磷酸(IP3)和钙离子,可以通过缝隙连接在细胞间进行传递。[17] 不同种类的缝隙连接也对不同的分子有不同的选择通透性。
- 一般来说,小于485道尔顿的分子可进行跨膜运动 [17] (1100道尔顿的分子则是通过无脊椎动物缝隙连接)[18],尽管不同的连接蛋白亚单位可能赋予不同的孔径和不同的电荷选择性。大的生物分子,例如核酸和蛋白质,是无法通过缝隙连接蛋白通道在细胞间进行胞质转移。
- 确保通过缝隙连接的分子和电流不会泄漏到细胞间隙中。
迄今为止,缝隙连接蛋白被发现有五种不同的功能:
- 细胞间的电和代谢耦合
- 通过半通道进行电和代谢交换
- 肿瘤抑制基因(Cx43、Cx32和Cx36)
- 与导电缝隙连接通道无关的黏附功能(神经在新皮层的迁移)
- 羧基末端在细胞质信号通路(Cx43)中的作用
4 发生和分布编辑
已经在细胞相互接触的各种动物器官和组织中均观察到缝隙连接的存在。从20世纪50年代到70年代,在小龙虾神经[19] 、大鼠胰腺、肝脏、肾上腺皮质、附睾、十二指肠、肌肉[20]、水蚤肝盲肠[21] 、水螅肌肉、猴视网膜[22]、兔角膜[23]、鱼胚盘[24] 、青蛙胚胎、兔卵巢[25]、再聚集细胞[26][27]、蟑螂血细胞胶囊[28]、兔皮肤[29]、鸡胚[30]、人胰岛中的郎格罕氏细胞[31],金鱼和仓鼠压力声传感-前庭受体[32] 、七鳃鳗和被囊类动物的心脏[33][34] 、鼠细精管[35] 、子宫肌层[36] 、晶状体[37] 和头足类动物消化道上皮细胞[38]中检测到了缝隙连接。自20世纪70年代以来,几乎在所有相互接触的动物细胞中都发现了缝隙连接。到了20世纪90年代,共聚焦显微镜等新技术的出现可对大面积组织进行更快速的观察测量。自20世纪70年代以来,即使在传统被认为可能相互分离细胞的组织,如骨骼,也显示出缝隙连接的存在,即使十分微弱的存在。[39] 缝隙连接似乎存在于所有的动物器官和组织中,除了通常不与邻近细胞接触的细胞之外,而这种例外情况的发现显得十分有趣。成人骨骼肌可能就是是一个例外。有人可能会认为,如果存在于骨骼肌中,缝隙连接可能会以任意方式在构成肌肉的细胞中传播收缩行为。至少在某些情况下,这可能不像其他肌肉类型细胞存在缝隙连接。[40]通过对癌症或衰老过程的研究分析,[41][42][43] 可以发现缝隙连接减少或缺失的所导致的结果。[44]
5 功能编辑
缝隙连接可被看作是电信号、小分子和离子在细胞间的直接通路。如下所述,对这种通讯的控制可对多细胞生物产生复杂的下游效应。
5.1 胚胎、器官和组织发育
在20世纪80年代,人们研究了缝隙连接通信中更微妙且重要的作用。研究发现,在胚胎细胞中加入抗连接蛋白抗体可以破坏缝隙连接的物质交流。[45][46]导致具有封闭缝隙连接区域的胚胎无法正常发育。虽然抗体阻断缝隙连接的机制尚不清楚,但已经进行了系统研究来阐明这一机制。[47][48] 这些进一步的研究表明,缝隙连接似乎是动物细胞极性和左右对称/不对称发展的关键因子。 [49][50] 虽然决定身体器官位置的信号似乎依赖于缝隙连接,但胚胎发育后期细胞更基本的分化也是如此。[51][52][53][54][55]此外,缝隙连接也被发现具有负责传输药物产生效果所需的信号的功能,相反,一些药物被证明可以阻断缝隙连接通道。[56]
5.2 缝隙连接和“旁观者效应”
细胞死亡
“旁观者效应”的含义是无辜的旁观者被杀害,它也是由缝隙连接介导的。当细胞因疾病或损伤而受到影响并开始死亡时,信息通过缝隙连接传输到与死亡细胞相连的邻近细胞。这可能会导致原本未受影响的健康旁观细胞也发生死亡。[57] 因此,在患病细胞中,考虑旁观者效应至关重要,这为获得更多资金和蓬勃发展的研究开辟途径。[58][59][60][61][62][63][64][65][66] 后来,旁观者效应也被用于研究辐射或机械力所损伤的细胞,以及因此所引起的伤口愈合过程中。[67][68][69][70][71] 而疾病似乎也会影响缝隙连接在伤口愈合中发挥的功能。[72][73]
组织重构
尽管由于潜在的治疗可能性,人们倾向于关注疾病中的旁观者效应,但证据表明,在组织的正常发育中,旁观者效应起着更为重要的作用。一些细胞及其周围基质的死亡可能是组织达到起最终形态的必要条件,缝隙连接在这一过程也显得十分关键。[74][75]在更复杂研究中,试图将我们对缝隙连接在伤口愈合和组织发育中的同时所发挥的作用的理解结合起来。[76][77] [78]
5.3 电气耦合领域
在大多数动物体内的细胞里,缝隙连接在电性和化学性上进行耦合。电耦合可以相对快速地发挥作用。本节中提及的组织(以及器官和系统)具有众所周知的功能,观察到这些功能由缝隙连接协调,细胞间信号发生在几微秒或更短的时间范围内。
心脏
缝隙连接在心肌中特别重要:收缩信号通过缝隙连接有效传递,使心肌细胞能够一致性收缩。缝隙连接几乎在身体的所有组织中都有表达,除了成年人发育完全的骨骼肌和可移动细胞类型,如精子或红细胞。一些人类遗传疾病与缝隙连接基因突变相关。其中许多影响了皮肤组织,因为这种组织严重依赖于缝隙连接通讯来调节其分化和增殖。强心剂可以在药理学上打开心脏缝隙连接。
神经元
位于神经元中的缝隙连接通常被称为电突触。在缝隙连接结构被发现之前,通过电测量发现了电突触。电突触存在于整个中枢神经系统之中,并且已经在脊椎动物的新皮质、海马、前庭核、丘脑网状核、锁膜下核、下橄榄核、三叉神经中脑核、腹侧被盖区、嗅球、视网膜和脊髓中进行了专门研究。[79]
在锁膜下核、小脑、浦肯野神经元和博格曼神经胶质细胞之间存在弱神经元与神经胶质细胞耦合。星形胶质细胞与其他星形胶质细胞以及少突胶质细胞通过缝隙链接进行偶联。[80]此外,缝隙连接基因Cx43和Cx56.6的突变会导致脑白质退化,与髓鞘蛋白编码基因缺陷病和多发性硬化症相似。
在神经元缝隙连接中表达的连接蛋白包括:
- mCX36
- mCX57
- mCX45
检测到至少五种其他连接蛋白(mCx26、mCx30.2、mCx32、mCx43、mCx47)的mRNA,但在超微结构定义的缝隙连接中没有发现相应蛋白的免疫细胞化学证据。这些mRNA似乎被细胞类型和细胞谱系特异性的miRNA所下调或破坏。
视网膜
视网膜内的神经元存在广泛的耦合,缝隙连接存在于无论是在同一种细胞类型的群体内,还是在不同细胞类型之间。[81]
6 探索编辑
6.1 命名
缝隙连接之所以如此命名,是因为在两个细胞之间的这些特殊连接处存在“间隙”。[82] 随着透射电子显微镜(TEM)分辨率的提高,在1953年左右缝隙结构首次被观察和描述。
“缝隙连接”这个术语出现在大约16年后的1969年左右。[83][84][85] 当时用透射电镜拍摄的其他细胞间连接并未显示出类似的狭窄规则间隙。
6.2 形成功能指标
远在缝隙连接中的“缝隙”出现之前,它们就出现在相邻神经细胞的交界处。缝隙连接处相邻细胞膜的紧密接触,使得研究人员推测它们在细胞间的通讯,特别是电信号的传输中发挥了作用。[86][87][88] 缝隙连接也被证明是电整流的,被称为电突触。[89][90]后来发现,化学物质也可以通过缝隙连接在细胞间传输。[91]
在大多数早期研究中,无论是隐式或显式的,缝隙连接的面积在结构上不同于周围的膜,使其看起来有所不同。缝隙连接被证明在细胞外空间或“缝隙”中的两个细胞之间产生了微环境。这部分细胞外空间与周围空间有些隔离,也由我们现在所提到的连接子对桥接起来,连接子对形成更紧密的桥,跨越两个细胞之间的缝隙连接缝隙。当通过冷冻断裂技术在膜平面上观察时,实现缝隙连接斑块内连接子的更高分辨率分布观察是可能的。[92]
连接蛋白游离岛在一些连接处被观察到。在帕拉契亚(Peracchia)借助透射电镜显示囊泡与缝隙连接斑块系统相关之前,基本上未有相关的实验研究解释。[93]帕拉契亚( Peracchia)的工作可能也是第一个描述成对连接结构的研究,他称之为“球状体”。显示囊泡与缝隙连接相关,并提出囊泡内容物可能穿过两个细胞之间的连接斑块,此类的研究很少,因为大多数研究集中于连接子而不是囊泡。后来一项结合显微镜技术的研究证实了细胞间囊泡转移中缝隙连接发挥功能的早期证据。囊泡转移区域与缝隙连接斑块内的连接蛋白游离岛相关。[94]
6.3 电和化学神经突触
由于在神经细胞以外的细胞类型中缝隙连接广泛存在,“缝隙连接”一词变得比其他术语更加普遍,如电突触或连接。通过研究化学突触的形成和缝隙连接的存在,揭示了神经细胞和缝隙连接之间关系的另一个维度。通过追踪水蛭的缝隙连接表达被抑制的神经发育,表明双向缝隙连接(电神经突触)需要在两个细胞之间形成,然后才能生长形成一个单向的“化学神经突触”。[95] 化学神经突触常被称为突触,是对更模糊术语“神经突触”的简称。
6.4 构成
连接蛋白
纯化[96][97] 富集于通道形成蛋白(连接蛋白)的细胞缝隙连接斑块,在x射线衍射下显示出形成六角形阵列的形态。现在,系统研究和鉴定[98]主要的缝隙连接蛋白成为可能 。透射电镜的精细超微结构显示,[99][100] 在参与缝隙连接斑块的两个细胞中,蛋白质以互补的方式出现。缝隙连接斑块是在透射电镜薄层中观察到的相对较大的膜面积,通过冷冻断裂(FF)以更温和的处理条件下,观察到在两种组织的缝隙连接中充满跨膜蛋白。由于仅一种蛋白质就能在缝隙链接中实现细胞间通讯,[101] 缝隙连接一词往往成为一组组装连接蛋白的同义词,尽管这些现象并未在体内显现出来。对来自各种组织的富含缝隙连接分离物的生化分析表明,存在一个连接蛋白家族。[102][103][104]
已报道的分离缝隙连接的超微结构和生物化学表明,缝隙连接蛋白在缝隙连接斑块或结构域中优先出现,而缝隙连接蛋白是其中最具特征的成分。已经注意到,将蛋白质组织成具有缝隙连接斑块的阵列可能具有重要意义。[105][105]此项早期工作很可能已经反映了缝隙连接中存在的不仅仅是连接蛋白。结合新近出现的冷冻断裂内膜观察和免疫细胞化学标记细胞成分的方法显示(冷冻断裂复制免疫标记或FRIL和薄切片免疫标记),体内缝隙连接斑块含有连接蛋白。[106][107] 最近,使用免疫荧光显微镜对更大面积组织的研究,澄清了早期结果中的预示的多样性。缝隙连接斑块被证实具有组成可变性,是连接蛋白和非连接蛋白的家,也使得术语“缝隙连接”和“缝隙连接斑块”的现代用法不可互换。[108] 换句话说,常用术语“缝隙连接”是指包含连接蛋白的结构,而缝隙连接斑块也可能包含其他结构特征的定义。
“斑块”或“形成斑块”
早期对“缝隙连接”和“缝隙连接”的描述并没有这样称呼它们,而是使用了许多其他术语。“突触盘”[109]很可能是缝隙连接斑块的准确称谓。虽然当时对连接体的详细结构和功能的描述有限,但总的“圆盘”结构相对较大,各种透射电镜技术使其很容易观察到。研究人员能够利用透射电镜在体内和体外轻松定位包含在盘状斑块内的连接子。圆盘或“斑块”似乎具有不同于连接蛋白传递的结构性质。[110] 人们认为,如果斑块中的膜区域具有传输信号的能力,膜区域必须以某种方式密封,以防止泄漏。[110]后来的研究表明,缝隙连接斑块是非连接蛋白的家,使得“缝隙连接”和“缝隙连接斑块”这两个术语的现代用法不可互换,因为缝隙连接斑块的区域可能含有连接蛋白以外的其它蛋白。[108][111] 正如连接蛋白不总是占据斑块的整个区域,文献中描述的其他成分可能只是以长期或短期形式存在。[112]
观察缝隙连接膜平面内的研究表明,在连接蛋白进入细胞之前,两个细胞之间形成了“斑块形成”。透射电镜和冷冻断裂薄层观察时,它们是颗粒自由区域,表明可能他们非常小或不存在跨膜蛋白质。我们对斑块形成的结构知之甚少,也不知道当连接蛋白和其他成分进入或离开时,形成斑块的结构是怎样变化的。早期关于小缝隙连接形成的研究之一,描述了成排的粒子和自由粒子环。[113]随着缝隙连接增大,它们被描述为连接蛋白进入时所形成斑块。颗粒缝隙连接被认为在形成斑块后4-6小时出现的。[114] 如何利用微管蛋白将连接蛋白转运至斑块也变得越来越清楚。[115][115]
早期研究者很难分析经典缝隙连接斑块的形成斑块和非连接蛋白部分。透射电镜荧光显微镜下和冷冻断层图像显示其似乎是一个脂膜结构域,可以对其他脂质和蛋白质形成相对刚性的屏障。有间接证据表明某些脂质优先参与形成斑块,但这不能被认为是决定性的。[116][117] 很难设想在不影响膜斑块组成的情况下,将膜拆开来分析膜斑块。通过对膜中连接蛋白的研究,研究了与连接蛋白相关的脂质。[118] 研究发现,特异性连接蛋白倾向于优先与特定磷脂结合。由于形成斑块先于连接蛋白形成,这些结果仍然不能确定斑块本身组成的独特之处。其他研究结果显示,连接蛋白与另一连接处使用的蛋白支架相关,即闭锁小带ZO1。[119] 虽然这有助于我们理解连接蛋白是如何进入缝隙连接形成斑块的,但斑块本身的组成的理解仍然较为粗略。利用透射电镜FRIL在缝隙连接斑块的体内成分的研究方面取得了一些进展。[112][119]
参考文献
- [1]
^White, Thomas W.; Paul, David L. (1999). "Genetic diseases and gene knockouts reveal diverse connexin functions". Annual Review of Physiology. 61 (1): 283–310. doi:10.1146/annurev.physiol.61.1.283. PMID 10099690..
- [2]
^Kelsell, David P.; Dunlop, John; Hodgins, Malcolm B. (2001). "Human diseases: clues to cracking the connexin code?". Trends in Cell Biology. 11 (1): 2–6. doi:10.1016/S0962-8924(00)01866-3. PMID 11146276..
- [3]
^Willecke, Klaus; Eiberger, Jürgen; Degen, Joachim; Eckardt, Dominik; Romualdi, Alessandro; Güldenagel, Martin; Deutsch, Urban; Söhl, Goran (2002). "Structural and functional diversity of connexin genes in the mouse and human genome". Biological Chemistry. 383 (5): 725–37. doi:10.1515/BC.2002.076. PMID 12108537..
- [4]
^Lampe, Paul D.; Lau, Alan F. (2004). "The effects of connexin phosphorylation on gap junctional communication". The International Journal of Biochemistry & Cell Biology. 36 (7): 1171–86. doi:10.1016/S1357-2725(03)00264-4. PMC 2878204. PMID 15109565..
- [5]
^Lampe, Paul D.; Lau, Alan F. (2000). "Regulation of gap junctions by phosphorylation of connexins". Archives of Biochemistry and Biophysics. 384 (2): 205–15. doi:10.1006/abbi.2000.2131. PMID 11368307..
- [6]
^C. elegans Sequencing, Consortium (Dec 11, 1998). "Genome sequence of the nematode C. elegans: a platform for investigating biology". Science. 282 (5396): 2012–8. doi:10.1126/science.282.5396.2012. PMID 9851916..
- [7]
^Ganfornina, MD; Sánchez, D; Herrera, M; Bastiani, MJ (1999). "Developmental expression and molecular characterization of two gap junction channel proteins expressed during embryogenesis in the grasshopper Schistocerca americana". Developmental Genetics. 24 (1–2): 137–50. doi:10.1002/(SICI)1520-6408(1999)24:1/2<137::AID-DVG13>3.0.CO;2-7. hdl:10261/122956. PMID 10079517..
- [8]
^Starich, T. A. (1996). "eat-5 and unc-7 represent a multigene family in Caenorhabditis elegans involved in cell-cell coupling". J. Cell Biol. 134 (2): 537–548. doi:10.1083/jcb.134.2.537. PMC 2120886. PMID 8707836..
- [9]
^Simonsen, Karina T.; Moerman, Donald G.; Naus, Christian C. (2014). "Gap junctions in C. elegans". Frontiers in Physiology. 5: 40. doi:10.3389/fphys.2014.00040. PMC 3920094. PMID 24575048..
- [10]
^Barbe, M. T. (1 April 2006). "Cell-Cell Communication Beyond Connexins: The Pannexin Channels". Physiology. 21 (2): 103–114. doi:10.1152/physiol.00048.2005. PMID 16565476..
- [11]
^Panchina, Yuri; Kelmanson, Ilya; Matz, Mikhail; Lukyanov, Konstantin; Usman, Natalia; Lukyanov, Sergey (June 2000). "A ubiquitous family of putative gap junction molecules". Current Biology. 10 (13): R473–R474. Bibcode:1996CBio....6.1213A. doi:10.1016/S0960-9822(00)00576-5. PMID 10898987..
- [12]
^Lohman, Alexander W.; Isakson, Brant E. (2014). "Differentiating connexin hemichannels and pannexin channels in cellular ATP release". FEBS Letters. 588 (8): 1379–1388. doi:10.1016/j.febslet.2014.02.004. PMC 3996918. PMID 24548565..
- [13]
^Maeda, Shoji; Nakagawa, So; Suga, Michihiro; Yamashita, Eiki; Oshima, Atsunori; Fujiyoshi, Yoshinori; Tsukihara, Tomitake (2009). "Structure of the connexin 26 gap junction channel at 3.5 A resolution". Nature. 458 (7238): 597–602. Bibcode:2009Natur.458..597M. doi:10.1038/nature07869. PMID 19340074..
- [14]
^Hsieh, CL; Kumar, NM; Gilula, NB; Francke, U (Mar 1991). "Distribution of genes for gap junction membrane channel proteins on human and mouse chromosomes". Somatic Cell and Molecular Genetics. 17 (2): 191–200. doi:10.1007/bf01232976. PMID 1849321..
- [15]
^Kumar, NM; Gilula, NB (Feb 1992). "Molecular biology and genetics of gap junction channels". Seminars in Cell Biology. 3 (1): 3–16. doi:10.1016/s1043-4682(10)80003-0. PMID 1320430..
- [16]
^Kren, BT; Kumar, NM; Wang, SQ; Gilula, NB; Steer, CJ (Nov 1993). "Differential regulation of multiple gap junction transcripts and proteins during rat liver regeneration". The Journal of Cell Biology. 123 (3): 707–18. doi:10.1083/jcb.123.3.707. PMC 2200133. PMID 8227133..
- [17]
^Alberts, Bruce (2002). Molecular biology of the cell (4th ed.). New York: Garland Science. ISBN 978-0-8153-3218-3.[页码请求].
- [18]
^Loewenstein WR (July 1966). "Permeability of membrane junctions". Ann. N. Y. Acad. Sci. 137 (2): 441–72. Bibcode:1966NYASA.137..441L. doi:10.1111/j.1749-6632.1966.tb50175.x. PMID 5229810..
- [19]
^Robertson, JD (February 1953). "Ultrastructure of two invertebrate synapses". Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine. 82 (2): 219–23. doi:10.3181/00379727-82-20071. PMID 13037850..
- [20]
^Friend DS, Gilula NB (June 1972). "Variations in tight and gap junctions in mammalian tissues". J. Cell Biol. 53 (3): 758–76. doi:10.1083/jcb.53.3.758. PMC 2108762. PMID 4337577..
- [21]
^Hudspeth, AJ; Revel, JP. (Jul 1971). "Coexistence of gap and septate junctions in an invertebrate epithelium". J. Cell Biol. 50 (1): 92–101. doi:10.1083/jcb.50.1.92. PMC 2108432. PMID 5563454..
- [22]
^Raviola, E; Gilula, NB. (Jun 1973). "Gap junctions between photoreceptor cells in the vertebrate retina". Proc Natl Acad Sci U S A. 70 (6): 1677–81. Bibcode:1973PNAS...70.1677R. doi:10.1073/pnas.70.6.1677. PMC 433571. PMID 4198274..
- [23]
^Kreutziger GO (September 1976). "Lateral membrane morphology and gap junction structure in rabbit corneal endothelium". Exp. Eye Res. 23 (3): 285–93. doi:10.1016/0014-4835(76)90129-9. PMID 976372..
- [24]
^Lentz TL, Trinkaus JP (March 1971). "Differentiation of the junctional complex of surface cells in the developing Fundulus blastoderm". J. Cell Biol. 48 (3): 455–72. doi:10.1083/jcb.48.3.455. PMC 2108114. PMID 5545331..
- [25]
^Albertini, DF; Anderson, E. (Oct 1974). "The appearance and structure of intercellular connections during the ontogeny of the rabbit ovarian follicle with particular reference to gap junctions". J Cell Biol. 63 (1): 234–50. doi:10.1083/jcb.63.1.234. PMC 2109337. PMID 4417791..
- [26]
^Johnson R, Hammer M, Sheridan J, Revel JP (November 1974). "Gap junction formation between reaggregated Novikoff hepatoma cells". Proc. Natl. Acad. Sci. U.S.A. 71 (11): 4536–40. Bibcode:1974PNAS...71.4536J. doi:10.1073/pnas.71.11.4536. PMC 433922. PMID 4373716..
- [27]
^Knudsen, KA; Horwitz, AF. (1978). "Toward a mechanism of myoblast fusion". Prog Clin Biol Res. 23: 563–8. PMID 96453..
- [28]
^Baerwald RJ (1975). "Inverted gap and other cell junctions in cockroach hemocyte capsules: a thin section and freeze-fracture study". Tissue Cell. 7 (3): 575–85. doi:10.1016/0040-8166(75)90027-0. PMID 1179417..
- [29]
^Prutkin L (February 1975). "Mucous metaplasia and gap junctions in the vitamin A acid-treated skin tumor, keratoacanthoma". Cancer Res. 35 (2): 364–9. PMID 1109802..
- [30]
^Bellairs, R; Breathnach, AS; Gross, M. (Sep 1975). "Freeze-fracture replication of junctional complexes in unincubated and incubated chick embryos". Cell Tissue Res. 162 (2): 235–52. doi:10.1007/BF00209209. PMID 1237352..
- [31]
^Orci L, Malaisse-Lagae F, Amherdt M, et al. (November 1975). "Cell contacts in human islets of Langerhans". J. Clin. Endocrinol. Metab. 41 (5): 841–4. doi:10.1210/jcem-41-5-841. PMID 1102552..
- [32]
^Hama K, Saito K (February 1977). "Gap junctions between the supporting cells in some acoustico-vestibular receptors". J. Neurocytol. 6 (1): 1–12. doi:10.1007/BF01175410. PMID 839246..
- [33]
^Shibata, Y; Yamamoto, T (March 1977). "Gap junctions in the cardiac muscle cells of the lamprey". Cell Tissue Res. 178 (4): 477–82. doi:10.1007/BF00219569. PMID 870202..
- [34]
^Lorber, V; Rayns, DG (April 1977). "Fine structure of the gap junction in the tunicate heart". Cell Tissue Res. 179 (2): 169–75. doi:10.1007/BF00219794. PMID 858161..
- [35]
^McGinley D, Posalaky Z, Provaznik M (October 1977). "Intercellular junctional complexes of the rat seminiferous tubules: a freeze-fracture study". Anat. Rec. 189 (2): 211–31. doi:10.1002/ar.1091890208. PMID 911045..
- [36]
^Garfield, RE; Sims, SM; Kannan, MS; Daniel, EE (November 1978). "Possible role of gap junctions in activation of myometrium during parturition". Am. J. Physiol. 235 (5): C168–79. doi:10.1152/ajpcell.1978.235.5.C168. PMID 727239..
- [37]
^Goodenough, DA (November 1979). "Lens gap junctions: a structural hypothesis for nonregulated low-resistance intercellular pathways". Invest. Ophthalmol. Vis. Sci. 18 (11): 1104–22. PMID 511455..
- [38]
^Boucaud-Camou, Eve (1980). "Junctional structures in digestive epithelia of a cephalopod". Tissue Cell. 12 (2): 395–404. doi:10.1016/0040-8166(80)90013-0. PMID 7414602..
- [39]
^Jones SJ, Gray C, Sakamaki H, et al. (April 1993). "The incidence and size of gap junctions between the bone cells in rat calvaria". Anat. Embryol. 187 (4): 343–52. doi:10.1007/BF00185892. PMID 8390141..
- [40]
^Sperelakis, Nicholas; Ramasamy, Lakshminarayanan (2005). "Gap-junction channels inhibit transverse propagation in cardiac muscle". Biomed Eng Online. 4 (1): 7. doi:10.1186/1475-925X-4-7. PMC 549032. PMID 15679888..
- [41]
^Larsen WJ, Azarnia R, Loewenstein WR (June 1977). "Intercellular communication and tissue growth: IX. Junctional membrane structure of hybrids between communication-competent and communication-incompetent cells". J. Membr. Biol. 34 (1): 39–54. doi:10.1007/BF01870292. PMID 561191..
- [42]
^Corsaro CM, Migeon BR (October 1977). "Comparison of contact-mediated communication in normal and transformed human cells in culture". Proc. Natl. Acad. Sci. U.S.A. 74 (10): 4476–80. Bibcode:1977PNAS...74.4476C. doi:10.1073/pnas.74.10.4476. PMC 431966. PMID 270694..
- [43]
^Habermann, H; Chang, WY; Birch, L; Mehta, P; Prins, GS (January 2001). "Developmental exposure to estrogens alters epithelial cell adhesion and gap junction proteins in the adult rat prostate". Endocrinology. 142 (1): 359–69. doi:10.1210/en.142.1.359. PMID 11145599..
- [44]
^Kelley, Robert O.; Vogel, Kathryn G.; Crissman, Harry A.; Lujan, Christopher J.; Skipper, Betty E. (March 1979). "Development of the aging cell surface. Reduction of gap junction-mediated metabolic cooperation with progressive subcultivation of human embryo fibroblasts (IMR-90)". Exp. Cell Res. 119 (1): 127–43. doi:10.1016/0014-4827(79)90342-2. PMID 761600..
- [45]
^Warner, Anne E.; Guthrie, Sarah C.; Gilula, Norton B. (1984). "Antibodies to gap-junctional protein selectively disrupt junctional communication in the early amphibian embryo". Nature. 311 (5982): 127–31. Bibcode:1984Natur.311..127W. doi:10.1038/311127a0. PMID 6088995..
- [46]
^Warner, AE (1987). "The use of antibodies to gap junction protein to explore the role of gap junctional communication during development". Ciba Found. Symp. 125: 154–67. PMID 3030673..
- [47]
^Bastide, B; Jarry-Guichard, T; Briand, JP; Délèze, J; Gros, D (April 1996). "Effect of antipeptide antibodies directed against three domains of connexin43 on the gap junctional permeability of cultured heart cells". J. Membr. Biol. 150 (3): 243–53. doi:10.1007/s002329900048. PMID 8661989..
- [48]
^Hofer, A; Dermietzel, R (September 1998). "Visualization and functional blocking of gap junction hemichannels (connexons) with antibodies against external loop domains in astrocytes". Glia. 24 (1): 141–54. doi:10.1002/(SICI)1098-1136(199809)24:1<141::AID-GLIA13>3.0.CO;2-R. PMID 9700496..
- [49]
^Levin, Michael; Mercola, Mark (November 1998). "Gap junctions are involved in the early generation of left-right asymmetry". Dev. Biol. 203 (1): 90–105. doi:10.1006/dbio.1998.9024. PMID 9806775..
- [50]
^Levin, M; Mercola, M (November 1999). "Gap junction-mediated transfer of left-right patterning signals in the early chick blastoderm is upstream of Shh asymmetry in the node". Development. 126 (21): 4703–14. PMID 10518488..
- [51]
^Bani-Yaghoub, Mahmud; Underhill, T. Michael; Naus, Christian C.G. (1999). "Gap junction blockage interferes with neuronal and astroglial differentiation of mouse P19 embryonal carcinoma cells". Dev. Genet. 24 (1–2): 69–81. doi:10.1002/(SICI)1520-6408(1999)24:1/2<69::AID-DVG8>3.0.CO;2-M. PMID 10079512..
- [52]
^Bani-Yaghoub, Mahmud; Bechberger, John F.; Underhill, T.Michael; Naus, Christian C.G. (March 1999). "The effects of gap junction blockage on neuronal differentiation of human NTera2/clone D1 cells". Exp. Neurol. 156 (1): 16–32. doi:10.1006/exnr.1998.6950. PMID 10192774..
- [53]
^Donahue, HJ; Li, Z; Zhou, Z; Yellowley, CE (February 2000). "Differentiation of human fetal osteoblastic cells and gap junctional intercellular communication". Am. J. Physiol., Cell Physiol. 278 (2): C315–22. doi:10.1152/ajpcell.2000.278.2.C315. PMID 10666026..
- [54]
^Cronier, L.; Frendo, JL; Defamie, N; Pidoux, G; Bertin, G; Guibourdenche, J; Pointis, G; Malassine, A (November 2003). "Requirement of gap junctional intercellular communication for human villous trophoblast differentiation". Biol. Reprod. 69 (5): 1472–80. doi:10.1095/biolreprod.103.016360. PMID 12826585..
- [55]
^El-Sabban, M. E.; Sfeir, AJ; Daher, MH; Kalaany, NY; Bassam, RA; Talhouk, RS (September 2003). "ECM-induced gap junctional communication enhances mammary epithelial cell differentiation". J. Cell Sci. 116 (Pt 17): 3531–41. doi:10.1242/jcs.00656. PMID 12893812..
- [56]
^Srinivas, M.; Hopperstad, MG; Spray, DC (September 2001). "Quinine blocks specific gap junction channel subtypes". Proc. Natl. Acad. Sci. U.S.A. 98 (19): 10942–7. Bibcode:2001PNAS...9810942S. doi:10.1073/pnas.191206198. PMC 58578. PMID 11535816..
- [57]
^Li Bi, Wan; Parysek, Linda M.; Warnick, Ronald; Stambrook, Peter J. (December 1993). "In vitro evidence that metabolic cooperation is responsible for the bystander effect observed with HSV tk retroviral gene therapy". Hum. Gene Ther. 4 (6): 725–31. doi:10.1089/hum.1993.4.6-725. PMID 8186287..
- [58]
^Pitts, JD (November 1994). "Cancer gene therapy: a bystander effect using the gap junctional pathway". Mol. Carcinog. 11 (3): 127–30. doi:10.1002/mc.2940110302. PMID 7945800..
- [59]
^Colombo, Bruno M.; Benedetti, Sara; Ottolenghi, Sergio; Mora, Marina; Pollo, Bianca; Poli, Giorgio; Finocchiaro, Gaetano (June 1995). "The "bystander effect": association of U-87 cell death with ganciclovir-mediated apoptosis of nearby cells and lack of effect in athymic mice". Hum. Gene Ther. 6 (6): 763–72. doi:10.1089/hum.1995.6.6-763. PMID 7548276..
- [60]
^Fick, James; Barker, Fred G.; Dazin, Paul; Westphale, Eileen M.; Beyer, Eric C.; Israel, Mark A. (November 1995). "The extent of heterocellular communication mediated by gap junctions is predictive of bystander tumor cytotoxicity in vitro". Proc. Natl. Acad. Sci. U.S.A. 92 (24): 11071–5. Bibcode:1995PNAS...9211071F. doi:10.1073/pnas.92.24.11071. PMC 40573. PMID 7479939..
- [61]
^Elshami, AA; Saavedra, A; Zhang, H; Kucharczuk, JC; Spray, DC; Fishman, GI; Amin, KM; Kaiser, LR; Albelda, SM (January 1996). "Gap junctions play a role in the 'bystander effect' of the herpes simplex virus thymidine kinase/ganciclovir system in vitro". Gene Ther. 3 (1): 85–92. PMID 8929915..
- [62]
^Mesnil, Marc; Piccoli, Colette; Tiraby, Gerard; Willecke, Klaus; Yamasaki, Hiroshi (March 1996). "Bystander killing of cancer cells by herpes simplex virus thymidine kinase gene is mediated by connexins". Proceedings of the National Academy of Sciences of the United States of America. 93 (5): 1831–5. Bibcode:1996PNAS...93.1831M. doi:10.1073/pnas.93.5.1831. PMC 39867. PMID 8700844..
- [63]
^Shinoura, Nobusada; Chen, Lin; Wani, Maqsood A.; Kim, Young Gyu; Larson, Jeffrey J.; Warnick, Ronald E.; Simon, Matthias; Menon, Anil G.; et al. (May 1996). "Protein and messenger RNA expression of connexin43 in astrocytomas: implications in brain tumor gene therapy". J. Neurosurg. 84 (5): 839–45, discussion 846. doi:10.3171/jns.1996.84.5.0839. PMID 8622159..
- [64]
^Hamel, W; Magnelli, L; Chiarugi, VP; Israel, MA (June 1996). "Herpes simplex virus thymidine kinase/ganciclovir-mediated apoptotic death of bystander cells". Cancer Res. 56 (12): 2697–702. PMID 8665496..
- [65]
^Sacco, MG; Benedetti, S; Duflot-Dancer, A; Mesnil, M; Bagnasco, L; Strina, D; Fasolo, V; Villa, A; et al. (December 1996). "Partial regression, yet incomplete eradication of mammary tumors in transgenic mice by retrovirally mediated HSVtk transfer 'in vivo'". Gene Ther. 3 (12): 1151–6. PMID 8986442..
- [66]
^Ripps, Harris (March 2002). "Cell death in retinitis pigmentosa: gap junctions and the 'bystander' effect". Exp. Eye Res. 74 (3): 327–36. doi:10.1006/exer.2002.1155. PMID 12014914..
- [67]
^Little, JB; Azzam, EI; De Toledo, SM; Nagasawa, H (2002). "Bystander effects: intercellular transmission of radiation damage signals". Radiat Prot Dosimetry. 99 (1–4): 159–62. doi:10.1093/oxfordjournals.rpd.a006751. PMID 12194273..
- [68]
^Zhou, H; Randers-Pehrson, G; Suzuki, M; Waldren, CA; Hei, TK (2002). "Genotoxic damage in non-irradiated cells: contribution from the bystander effect". Radiat Prot Dosimetry. 99 (1–4): 227–32. doi:10.1093/oxfordjournals.rpd.a006769. PMID 12194291..
- [69]
^Lorimore, SA; Wright, EG (January 2003). "Radiation-induced genomic instability and bystander effects: related inflammatory-type responses to radiation-induced stress and injury? A review". Int. J. Radiat. Biol. 79 (1): 15–25. doi:10.1080/0955300021000045664. PMID 12556327..
- [70]
^Ehrlich, HP; Diez, T (2003). "Role for gap junctional intercellular communications in wound repair". Wound Repair Regen. 11 (6): 481–9. doi:10.1046/j.1524-475X.2003.11616.x. PMID 14617290..
- [71]
^Coutinho, P.; Qiu, C.; Frank, S.; Wang, C.M.; Brown, T.; Green, C.R.; Becker, D.L. (July 2005). "Limiting burn extension by transient inhibition of Connexin43 expression at the site of injury". Br J Plast Surg. 58 (5): 658–67. doi:10.1016/j.bjps.2004.12.022. PMID 15927148..
- [72]
^Wang, C. M.; Lincoln, J.; Cook, J. E.; Becker, D. L. (November 2007). "Abnormal connexin expression underlies delayed wound healing in diabetic skin". Diabetes. 56 (11): 2809–17. doi:10.2337/db07-0613. PMID 17717278..
- [73]
^Rivera, EM; Vargas, M; Ricks-Williamson, L (1997). "Considerations for the aesthetic restoration of endodontically treated anterior teeth following intracoronal bleaching". Pract Periodontics Aesthet Dent. 9 (1): 117–28. PMID 9550065..
- [74]
^Cusato, K; Bosco, A; Rozental, R; Guimarães, CA; Reese, BE; Linden, R; Spray, DC (July 2003). "Gap junctions mediate bystander cell death in developing retina". J. Neurosci. 23 (16): 6413–22. doi:10.1523/JNEUROSCI.23-16-06413.2003. PMID 12878681..
- [75]
^Moyer, Kurtis E.; Saggers, Gregory C.; Ehrlich, H. Paul (2004). "Mast cells promote fibroblast populated collagen lattice contraction through gap junction intercellular communication". Wound Repair Regen. 12 (3): 269–75. doi:10.1111/j.1067-1927.2004.012310.x. PMID 15225205..
- [76]
^Djalilian, A. R.; McGaughey, D; Patel, S; Seo, EY; Yang, C; Cheng, J; Tomic, M; Sinha, S; et al. (May 2006). "Connexin 26 regulates epidermal barrier and wound remodeling and promotes psoriasiform response". J. Clin. Invest. 116 (5): 1243–53. doi:10.1172/JCI27186. PMC 1440704. PMID 16628254..
- [77]
^Zhang, Y.; Wang, H.; Kovacs, A.; Kanter, E. M.; Yamada, K. A. (February 2010). "Reduced expression of Cx43 attenuates ventricular remodeling after myocardial infarction via impaired TGF-beta signaling". Am. J. Physiol. Heart Circ. Physiol. 298 (2): H477–87. doi:10.1152/ajpheart.00806.2009. PMC 2822575. PMID 19966054..
- [78]
^Ey B, Eyking A, Gerken G, Podolsky DK, Cario E (August 2009). "TLR2 mediates gap junctional intercellular communication through connexin-43 in intestinal epithelial barrier injury". J. Biol. Chem. 284 (33): 22332–43. doi:10.1074/jbc.M901619200. PMC 2755956. PMID 19528242..
- [79]
^Connors; Long (2004). "Electrical synapses in the mammalian brain". Annu Rev Neurosci. 27: 393–418. doi:10.1146/annurev.neuro.26.041002.131128. PMID 15217338..
- [80]
^Orthmann-Murphy, Jennifer L.; Abrams, Charles K.; Scherer, Steven S. (May 2008). "Gap Junctions Couple Astrocytes and Oligodendrocytes". Journal of Molecular Neuroscience. 35 (1): 101–116. doi:10.1007/s12031-007-9027-5. PMC 2650399. PMID 18236012..
- [81]
^Béla Völgyi, Stewart A. Bloomfield (February 2009). "The diverse functional roles and regulation of neuronal gap junctions in the retina". Nature Reviews Neuroscience. 10 (7): 495–506. doi:10.1016/S0165-0173(99)00070-3. PMC 3381350. PMID 19491906..
- [82]
^Revel, J. P.; Karnovsky, M. J.; Aitchison, EJ; Smith, EG; Farrell, ID; Gutschik, E (1967). "Hexagonal array of subunits in intercellular junctions of the mouse heart and liver". The Journal of Cell Biology. 33 (3): C7–C12. doi:10.1083/jcb.33.3.C7. PMC 2107199. PMID 6036535..
- [83]
^Brightman, MW; Reese, TS (March 1969). "Junctions between intimately apposed cell membranes in the vertebrate brain". J. Cell Biol. 40 (3): 648–77. doi:10.1083/jcb.40.3.648. PMC 2107650. PMID 5765759..
- [84]
^Uehara Y, Burnstock G (January 1970). "Demonstration of "gap junctions" between smooth muscle cells". J. Cell Biol. 44 (1): 215–7. doi:10.1083/jcb.44.1.215. PMC 2107775. PMID 5409458..
- [85]
^Goodenough, DA; Revel, JP (May 1970). "A fine structural analysis of intercellular junctions in the mouse liver". J. Cell Biol. 45 (2): 272–90. doi:10.1083/jcb.45.2.272. PMC 2107902. PMID 4105112..
- [86]
^Robertson, J. D. (1953). "Ultrastructure of two invertebrate synapses". Proceedings of the Society for Experimental Biology and Medicine. 82 (2): 219–23. doi:10.3181/00379727-82-20071. PMID 13037850..
- [87]
^Robertson, J. D. (1963). Locke, Michael, ed. Cellular membranes in development. New York: Academic Press. OCLC 261587041.[页码请求].
- [88]
^Robertson (1981). "Membrane structure". The Journal of Cell Biology. 91 (3): 189s–204s. doi:10.1083/jcb.91.3.189s. JSTOR 1609517. PMC 2112820. PMID 7033238..
- [89]
^Furshpan, E. J.; Potter, D. D. (1957). "Mechanism of Nerve-Impulse Transmission at a Crayfish Synapse". Nature. 180 (4581): 342–3. Bibcode:1957Natur.180..342F. doi:10.1038/180342a0. PMID 13464833..
- [90]
^Furshpan; Potter, DD (1959). "Transmission at the giant motor synapses of the crayfish". The Journal of Physiology. 145 (2): 289–325. doi:10.1113/jphysiol.1959.sp006143. PMC 1356828. PMID 13642302..
- [91]
^Payton, B. W.; Bennett, M. V. L.; Pappas, G. D. (December 1969). "Permeability and structure of junctional membranes at an electrotonic synapse". Science. 166 (3913): 1641–3. Bibcode:1969Sci...166.1641P. doi:10.1126/science.166.3913.1641. PMID 5360587..
- [92]
^Chalcroft, J. P.; Bullivant, S (October 1970). "An interpretation of liver cell membrane and junction structure based on observation of freeze-fracture replicas of both sides of the fracture". J. Cell Biol. 47 (1): 49–60. doi:10.1083/jcb.47.1.49. PMC 2108397. PMID 4935338..
- [93]
^Peracchia, C (April 1973). "Low resistance junctions in crayfish. II. Structural details and further evidence for intercellular channels by freeze-fracture and negative staining". J. Cell Biol. 57 (1): 54–65. doi:10.1083/jcb.57.1.54. PMC 2108965. PMID 4120610..
- [94]
^Gruijters, W (2003). "Are gap junction membrane plaques implicated in intercellular vesicle transfer?". Cell Biol. Int. 27 (9): 711–7. doi:10.1016/S1065-6995(03)00140-9. PMID 12972275..
- [95]
^Todd KL, Kristan WB, French KA (November 2010). "Gap junction expression is required for normal chemical synapse formation". J. Neurosci. 30 (45): 15277–85. doi:10.1523/JNEUROSCI.2331-10.2010. PMC 3478946. PMID 21068332..
- [96]
^Goodenough, D. A.; Stoeckenius, W (1972). "The isolation of mouse hepatocyte gap junctions : Preliminary Chemical Characterization and X-Ray Diffraction". The Journal of Cell Biology. 54 (3): 646–56. doi:10.1083/jcb.54.3.646. PMC 2200277. PMID 4339819..
- [97]
^Goodenough, D. A. (1974). "Bulk isolation of mouse hepatocyte gap junctions : Characterization of the Principal Protein, Connexin". The Journal of Cell Biology. 61 (2): 557–63. doi:10.1083/jcb.61.2.557. PMC 2109294. PMID 4363961..
- [98]
^Kumar, N. M.; Gilula, NB (1986). "Cloning and characterization of human and rat liver cDNAs coding for a gap junction protein". The Journal of Cell Biology. 103 (3): 767–76. doi:10.1083/jcb.103.3.767. PMC 2114303. PMID 2875078..
- [99]
^McNutt NS, Weinstein RS (December 1970). "The ultrastructure of the nexus. A correlated thin-section and freeze-cleave study". J. Cell Biol. 47 (3): 666–88. doi:10.1083/jcb.47.3.666. PMC 2108148. PMID 5531667..
- [100]
^Chalcroft, J. P.; Bullivant, S (1970). "An interpretation of liver cell membrane and junction structure based on observation of freeze-fracture replicas of both sides of the fracture". The Journal of Cell Biology. 47 (1): 49–60. doi:10.1083/jcb.47.1.49. PMC 2108397. PMID 4935338..
- [101]
^Young; Cohn, ZA; Gilula, NB (1987). "Functional assembly of gap junction conductance in lipid bilayers: demonstration that the major 27 kd protein forms the junctional channel". Cell. 48 (5): 733–43. doi:10.1016/0092-8674(87)90071-7. PMID 3815522..
- [102]
^Nicholson; Gros, DB; Kent, SB; Hood, LE; Revel, JP (1985). "The Mr 28,000 gap junction proteins from rat heart and liver are different but related". The Journal of Biological Chemistry. 260 (11): 6514–7. PMID 2987225..
- [103]
^Beyer, E. C.; Paul, DL; Goodenough, DA (1987). "Connexin43: a protein from rat heart homologous to a gap junction protein from liver". The Journal of Cell Biology. 105 (6 Pt 1): 2621–9. doi:10.1083/jcb.105.6.2621. PMC 2114703. PMID 2826492..
- [104]
^Kistler, J; Kirkland, B; Bullivant, S (1985). "Identification of a 70,000-D protein in lens membrane junctional domains". The Journal of Cell Biology. 101 (1): 28–35. doi:10.1083/jcb.101.1.28. PMC 2113615. PMID 3891760..
- [105]
^J. Cell Biol. 1974 Jul;62(1) 32-47.Assembly of gap junctions during amphibian neurulation. Decker RS, Friend DS..
- [106]
^Gruijters, WTM; Kistler, J; Bullivant, S; Goodenough, DA (1987). "Immunolocalization of MP70 in lens fiber 16-17-nm intercellular junctions". The Journal of Cell Biology. 104 (3): 565–72. doi:10.1083/jcb.104.3.565. PMC 2114558. PMID 3818793..
- [107]
^Gruijters, WTM; Kistler, J; Bullivant, S (1987). "Formation, distribution and dissociation of intercellular junctions in the lens". Journal of Cell Science. 88 (3): 351–9. PMID 3448099..
- [108]
^Gruijters, WTM (1989). "A non-connexon protein (MIP) is involved in eye lens gap-junction formation". Journal of Cell Science. 93 (3): 509–13. PMID 2691517..
- [109]
^Robertson, JD (October 1963). "The occurrence of a subunit pattern in the unit membranes of club endings in mauthner cell synapses in goldfish brains". J. Cell Biol. 19 (1): 201–21. doi:10.1083/jcb.19.1.201. PMC 2106854. PMID 14069795..
- [110]
^Hand, AR; Gobel, S (February 1972). "The structural organization of the septate and gap junctions of Hydra". J. Cell Biol. 52 (2): 397–408. doi:10.1083/jcb.52.2.397. PMC 2108629. PMID 4109925..
- [111]
^Gruijters, WTM (2003). "Are gap junction membrane plaques implicated in intercellular vesicle transfer?". Cell Biology International. 27 (9): 711–7. doi:10.1016/S1065-6995(03)00140-9. PMID 12972275..
- [112]
^Ozato-Sakurai N, Fujita A, Fujimoto T (2011). Wong NS, ed. "The distribution of phosphatidylinositol 4,5-bisphosphate in acinar cells of rat pancreas revealed with the freeze-fracture replica labeling method". PLoS ONE. 6 (8): e23567. Bibcode:2011PLoSO...623567O. doi:10.1371/journal.pone.0023567. PMC 3156236. PMID 21858170..
- [113]
^Decker, RS; Friend, DS (July 1974). "Assembly of gap junctions during amphibian neurulation". J. Cell Biol. 62 (1): 32–47. doi:10.1083/jcb.62.1.32. PMC 2109180. PMID 4135001..
- [114]
^Decker, RS (June 1976). "Hormonal regulation of gap junction differentiation". J. Cell Biol. 69 (3): 669–85. doi:10.1083/jcb.69.3.669. PMC 2109697. PMID 1083855..
- [115]
^Francis R, Xu X, Park H, et al. (2011). Brandner JM, ed. "Connexin43 modulates cell polarity and directional cell migration by regulating microtubule dynamics". PLoS ONE. 6 (10): e26379. Bibcode:2011PLoSO...626379F. doi:10.1371/journal.pone.0026379. PMC 3194834. PMID 22022608..
- [116]
^Meyer, R; Malewicz, B; Baumann, WJ; Johnson, RG (June 1990). "Increased gap junction assembly between cultured cells upon cholesterol supplementation". J. Cell Sci. 96 (2): 231–8. PMID 1698798..
- [117]
^Johnson, R. G.; Reynhout, J. K.; Tenbroek, E. M.; Quade, B. J.; Yasumura, T.; Davidson, K. G. V.; Sheridan, J. D.; Rash, J. E. (January 2012). "Gap junction assembly: roles for the formation plaque and regulation by the C-terminus of connexin43". Mol. Biol. Cell. 23 (1): 71–86. doi:10.1091/mbc.E11-02-0141. PMC 3248906. PMID 22049024..
- [118]
^Locke, Darren; Harris, Andrew L (2009). "Connexin channels and phospholipids: association and modulation". BMC Biol. 7 (1): 52. doi:10.1186/1741-7007-7-52. PMC 2733891. PMID 19686581..
- [119]
^Li X, Kamasawa N, Ciolofan C, et al. (September 2008). "Connexin45-containing neuronal gap junctions in rodent retina also contain connexin36 in both apposing hemiplaques, forming bihomotypic gap junctions, with scaffolding contributed by zonula occludens-1". J. Neurosci. 28 (39): 9769–89. doi:10.1523/JNEUROSCI.2137-08.2008. PMC 2638127. PMID 18815262..
暂无